MAximising Therapeutic Utility in Rheumatoid Arthritis
Our Research
Our aim in MATURA is to develop tests to identify which treatments will work best for which patients with RA. We will test both the inflamed joint tissue (synovium) itself and peripheral blood to identify factors (biomarkers) that can be used to select the treatment most likely to be effective in individual patients. The biomarkers may be genetic changes, proteins, immune cells, other molecular changes or, perhaps more likely, a combination of these. Experts in statistics will combine the information generated and develop algorithms to help clinicians recommend the treatments most likely to work to patients. We will focus on developing tests to 4 drugs used routinely in patients with RA: methotrexate (MTX), anti-tumour necrosis factor (anti-TNF), rituximab (RTX) and tocilizumab (TOC). In only ~1 in 3 cases does each drug completely control the symptoms of RA so developing tests that better target the right drug to the right patient would dramatically improve the care of people with RA.
Work Stream 1 – Will analyse synovial markers in the PEAC dataset (http://www.peac-mrc.mds.qmul.ac.uk) and run the prospective randomised clinical trial, STRAP, to test the hypothesis that discrete cellular and molecular signatures in the synovial tissue (pathotypes) can be utilized to predict the response to existing biological therapies.
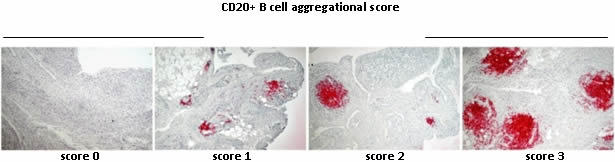
We will evaluate whether stratification of patients according to their synovial B cell infiltrate (poor or rich phenotypes) will better define response rates. We hypothesise that the Tocilizumab and Etanercept are superior to Rituximab in B-cell-poor patients. The ultimate aim is to provide a tailored approach to treatment decisions in patients at this stage of their disease, in order to maximise their potential response to therapy
Importantly, in collaboration with Industry Partners, we have drawn up plans to commercialise potential diagnostics for patient and economic benefit.
Work Stream 2 – We have already collected blood samples and clinical information from large groups of patients treated with each of the 3 drugs we are focusing on (data on ~3,000 anti-TNF, ~1,500 MTX and ~800 RTX treated patients). We have followed these patients from them starting the drug to 12 months later and, along the way, have collected blood samples, clinical information and data about the effectiveness and/or side effects of the drug.
We have consulted patients, through focus group interviews about their views and found them to be very supportive of precision medicine approaches.
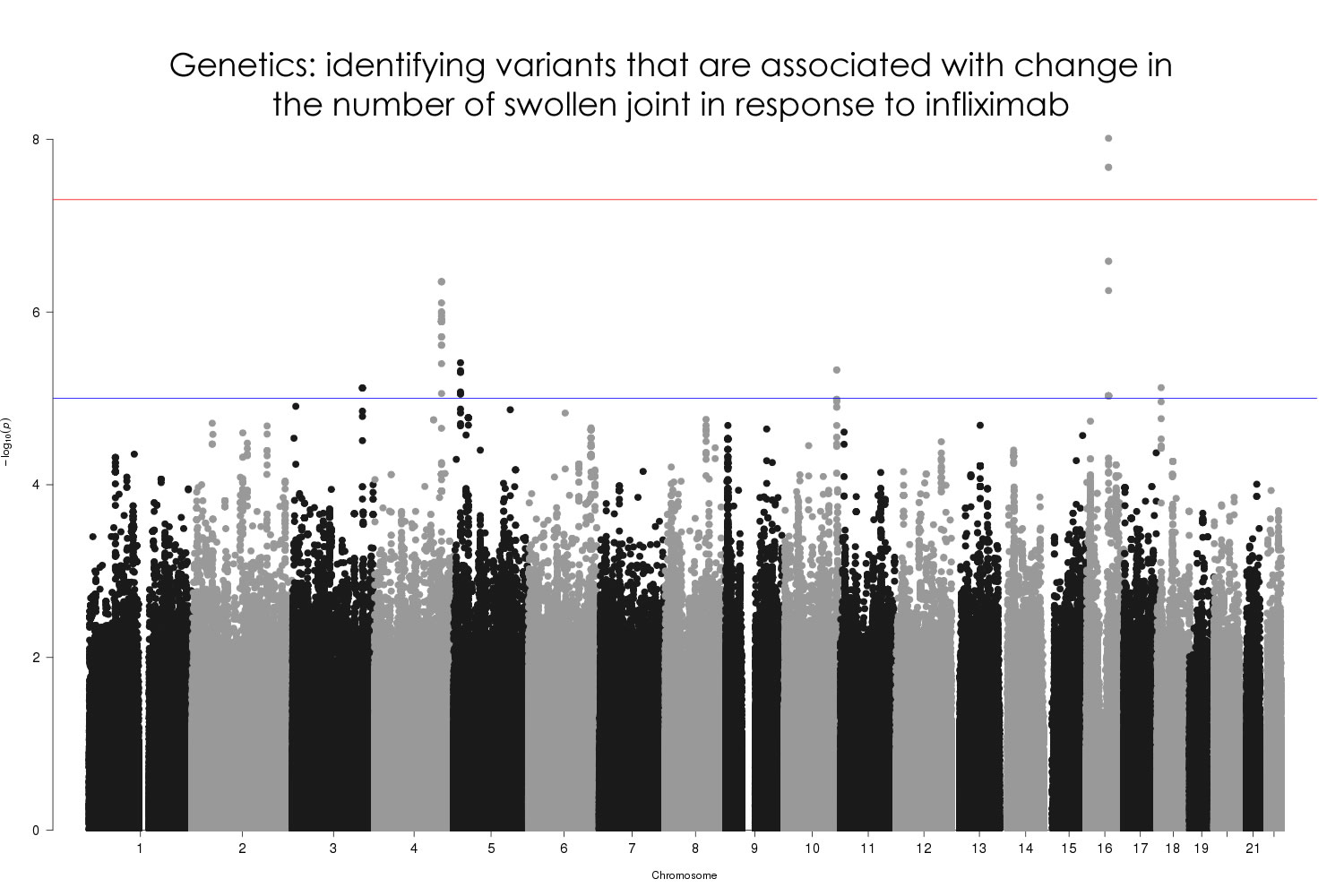
We have so far performed genome-wide SNP association analyses of response to MTX and anti-TNF in 1,424 and 1,752 RA patients respectively. In addition, we have taken a transcriptomics approach to understand outcomes in 246 patients (MTX=176, anti-TNF=70) and investigated genome-wide patterns of DNA methylation in 72 patients treated with MTX. MATURA is still in progress but analyses so far are indicating that, while falling short of clinical utility when taken in isolation, biomarkers of DAS28 response are detectable in whole-blood, correlate more strongly with the objective DAS28 components (for example the swollen joint count) and are observed in each of the genetic, transcriptomic and DNA methylation datasets.
The next phase of analysis will incorporate observed biomarkers, along with the relevant clinical data, in an integrative analysis to maximise the opportunity of detecting causal relationships, relevant biological pathways and predicting treatment outcomes for the major therapies used in RA.
The members of the MATURA consortium have collated all the planned analysis into a Data Analysis Plan, which is updated regularly as the programme develops (Please find latest Data Analysis Plan in Documentation area, BIOSTATS folder).
At present there is no genetic information that can reliably predict which patients will respond to therapies. Finding genetic predictors has been difficult and at present there are only two accepted genetic regions: CD84 and PDE3A-SLCO1C1. For this reason, we are now beginning to look at epigenetic factors; inheritable modifications that are not caused by sequence variation. We are particularly looking at methylation modifications of cytosine residues that are located in CpG sites (next to guanine residues). We are also looking at the differences in the levels of gene expression between patient groups with good response to therapy and those with a poor response. These data will then be integrated with existing genetic data to improve algorithms that will eventually enable patients to be stratified into treatment streams early in disease course.
The members of the MATURA consortium have collated all the planned analysis into a Data Analysis Plan, which is updated regularly as the programme develops (Please find latest Data Analysis Plan in Documentation area, BIOSTATS folder).
predictive of response to biological therapy in patients with rheumatoid arthritis